Inter-processor communication using the global array tools and the design of a data structure allowing distribution based on spacial decomposition are the key elements in taking advantage of the distribution of memory requirements and computational work with minimal communication.
In the spacial decomposition approach, the physical simulation volume is divided into rectangular boxes, each of which is assigned to a processor. Depending on the conditions of the calculation and the number of available processors, each processor contains one or more of these spacially grouped boxes. The most important aspects of this decomposition are the dependence of the box sizes and communication cost on the number of processors and the shape of the boxes, the frequent reassignment of atoms to boxes leading to a fluctuating number of atoms per box, and the locality of communication which is the main reason for the efficiency of this approach for very large molecular systems.
To improve efficiency, molecular systems are broken up into separately treated solvent and solute parts. Solvent molecules are assigned to the domains according to their center of geometry and are always owned by a one node. This avoids solvent-solvent bonded interactions crossing node boundaries. Solute molecules are broken up into segments, with each segment assigned to a processor based on its center of geometry. This limits the number of solute bonded interactions that cross node boundaries. The processor to which a particular box is assigned is responsible for the calculation of all interactions between atoms within that box. For the calculation of forces and energies in which atoms in boxes assigned to different processors are involved, data are exchanged between processors. The number of neighboring boxes is determined by the size and shape of the boxes and the range of interaction. The data exchange that takes place every simulation time step represents the main communication requirements. Consequently, one of the main efforts is to design algorithms and data structures to minimize the cost of this communication. However, for very large molecular systems, memory requirements also need to be taken into account.
To compromise between these requirements exchange of data is performed in successive point to point communications rather than using the shift algorithm which reduces the number of communication calls for the same amount of communicated data.
For inhomogeneous systems, the computational load of evaluating atomic interactions will generally differ between box pairs. This will lead to load imbalance between processors. Two algorithms have been implemented that allow for dynamically balancing the workload of each processor. One method is the dynamic resizing of boxes such that boxes gradually become smaller on the busiest node, thereby reducing the computational load of that node. Disadvantages of this method are that the efficiency depends on the solute distribution in the simulation volume and the redistribution of work depends on the number of nodes which could lead to results that depend on the number of nodes used. The second method is based on the dynamic redistribution of intra-node box-box interactions. This method represents a more coarse load balancing scheme, but does not have the disadvantages of the box resizing algorithm. For most molecular systems the box pair redistribution is the more efficient and preferred method.
The description of a molecular system consists of static and dynamic information. The static information does not change during a simulation and includes items such as connectivity, excluded and third neighbor lists, equilibrium values and force constants for all bonded and non-bonded interactions. The static information is called the topology of the molecular system, and is kept on a separate topology file. The dynamic information includes coordinates and velocities for all atoms in the molecular system, and is kept in a so-called restart file.
In molecular systems, a distinction is made between solvent and solute, which are treated separately. A solvent molecule is defined only once in the topology file, even though many solvent molecules usually are included in the actual molecular system. In the current implementation only one solvent can be defined. Everything that is not solvent in the molecular system is solute. Each solute atom in the system must be explicitly defined in the topology.
Molecules are defined in terms of one or more segments. Typically, repetitive parts of a molecule are each defined as a single segment, such as the amino acid residues in a protein. Segments can be quite complicated to define and are, therefore, collected in a set of database files. The definition of a molecular system in terms of segments is a sequence.
Topology files are created using the prepare module.
File names used have the form $system$_$calc$.$ext$, with
exception of the topology file (Section 27.1.2), which is named
$system$.top.
Anything that refers to the definition of the chemical system can be used
for $system$, as long as no periods or underlines are used.
The identifier $calc$ can be anything that refers to the type of
calculation to be performed for the system with the topology defined.
This file naming convention allows for the creation of a single
topology file $system$.top that can be used for a number of
different calculations, each identified with a different $calc$.
For example, if crown.top is the name of the topology file for
a crown ether, crown_em, crown_md, crown_ti could
be used with appropriate extensions for the filenames for energy
minimization, molecular dynamics simulation and multi-configuration
thermodynamic integration, respectively. All of these calculations
would use the same topology file crown.top.
The extensions <ext> identify the kind of information on a file,
and are pre-determined.
|
$ffield$_$level$,
$ffield$ is any of the
supported force fields (Section 27.1.5).
The source of the data is identified by $level$, and can be
| level | Description | Availability |
| s | original published data | public |
| x | additional published data | public |
| u | user preferred data | private |
| t | user defined temporary data | private |
Typically, only the level s and x databases are publicly
available.
The user is responsible for the private level u and t
database files. When the prepare module scans the databases, the priority
is t![]() u
u![]() x
x![]() s
s![]() .
.
The extension <ext> defines the type of database file within each
database directory.
|
The paths of the different database directories should be defined in a file .nwchemrc in a user's home directory, and provides the user the option to select which database files are scanned.
| Keyword | Force field |
| amber | AMBER95 |
| charmm | CHARMM |
Which atoms form a fragment is specified in the coordinate file, currently only in PDB format. the restriction is that bonded interactions may only involve atoms on at most two segments does no longer exist as of NWChem release 3.2.1. The segment entries define three sets of parameters for each interaction.
Free energy perturbations can be performed using set 1 for the
generation of the ensemble while using sets 2 and/or 3
as perturbations. Free energy multiconfiguration thermodynamic
integration and multistep thermodynamic perturbation calculations are
performed by gradually changing the interactions in the system from
parameter set 2 to parameter set 3. These modifications can be
edited into the segment files manually, or introduced directly into
the topology file using the modify commands in the input for
the prepare module.
The format of a segment is described in Tables 30.2-30.7.
The topology (Section 27.1.2) describes all static information that describes a molecular system. This includes the connectivity in terms of bond-stretching, angle-bending and torsional interactions, as well as the non-bonded van der Waals and Coulombic interactions.
The topology of a molecular system is generated by the prepare module from the sequence in terms of segments as specified on the PDB file. For each unique segment specified in this file the segment database directories are searched for the segment definition. For segments not found in one of the database directories a segment definition is generated in the temporary directory if a fragment file was found. If a fragment file could not be found, it is generated by the prepare module base on what is found on the PDB file.
When all segments are found or created, the parameter substitutions are
performed, using force field parameters taken from the parameter
databases. After all lists have been generated the
topology is written to a local topology file $system$.top.
Restart files contain all dynamical information about a molecular system and are created by the prepare module if a topology file is available. The prepare module will automatically generate coordinates for hydrogen atoms and monatomic counter ions not found on the PDB formatted coordinate file, if no fragment or segment files were generated using that PDB file.
The prepare module has a number of other optional input command, including solvation.
task md [ energy | optimize | dynamics | thermodynamics ]where the theory keyword md specifies use of the molecular dynamics module, and the operation keyword is one of
start
directive.
There is no default. If the rtdb name is given as system_calc,
the topology file used is system.top, while all other files
are named system_calc.ext.
set <integer iset>specifies the use of parameter set
<iset> for the
molecular dynamics simulation.
The topology file contains three separate parameters sets that can
be used. The default for <iset> is 1.
pset <integer isetp1> [<integer isetp2>]specifies the parameter sets to be used as perturbation potentials in single step thermodynamic perturbation free energy evaluations, where
<isetp1> specifies the first perturbation parameter set and
<isetp2> specifies the second perturbation parameter set. Legal
values for <isetp1> are 2 and 3. Legal value for <isetp2> is
3, in which case <isetp1> can only be 2. If specified, <iset>
is automatically set to 1.
pmfspecifies that any potential of mean force functions defined in the topology files are to be used.
distarspecifies that any distance restraint functions defined in the topology files are to be used.
sd <integer msdit> [init <real dx0sd>] [min <real dxsdmx>] \
[max <real dxmsd>]
specifies the variables for steepest descent energy minimizations,
where <msdit> is the maximum number of steepest descent steps taken,
for which the default is 100, <dx0sd> is the initial step size in nm
for which the default is 0.001, <dxsdmx> is the threshold for the
step size in nm for which the default is 0.0001, and <dxmsd> is the
maximum allowed step size in nm for which the default is 0.05.
cg <integer mcgit> [init <real dx0cg>] [min <real dxcgmx>] \
[cy <integer ncgcy>]
specifies the variables for conjugate gradient energy minimizations,
where <mcgit> is the maximum number of conjugate gradient steps
taken, for which the default is 100, <dx0cg> is the initial search
interval size in nm for which the default is 0.001, <dxcgmx> is the
threshold for the step size in nm for which the default is 0.0001, and
<ncgcy> is the number of conjugate gradient steps after which the
gradient history is discarded for which the default is 10. If conjugate
gradient energy minimization is preceded by steepest descent energy
minimization, the search interval is set to twice the final step of the
steepest descent energy minimization.
(forward | reverse) [[<integer mrun> of] <integer maxlam>]specifies the direction and number of integration steps in free energy evaluations, with forward being the default direction.
<mrun> is the number of ensembles that will be generated in
this calculation, and <maxlam> is the total number of ensembles
to complete the thermodynamic integration. The default value for
<maxlam> is 21. The default value of <mrun> is the
value of <maxlam>.
error <real edacq>specifies the maximum allowed statistical error in each generated ensemble, where
<edacq> is the maximum error allowed in the
ensemble average derivative of the Hamiltonian with respect to
drift <real ddacq>specifies the maximum allowed drift in the free energy result, where
<ddacq> is the maximum drift allowed in the
ensemble average derivative of the Hamiltonian with respect to
factor <real fdacq>specifies the maximum allowed change in ensemble size where
<fdacq> is the minimum size of an ensemble relative to the
previous ensemble in the calculation with a default value of 0.75.
decompspecifies that a free energy decomposition is to be carried out. Since free energy contributions are path dependent, results from a decomposition analysis can no be unambiguously interpreted, and the default is not to perform this decomposition.
sss [delta <real delta>]specifies that atomic non-bonded interactions describe a dummy atom in either the initial or final state of the thermodynamic calculation will be calculated using separation-shifted scaling, where
<delta>
is the separation-shifted scaling factor with a default of 0.075 nm
new | renew | extendspecifies the initial conditions for thermodynamic calculations. new indicates that this is an initial mcti calculation, which is the default. renew instructs to obtain the initial conditions for each
leapfrog | leapfrog_bcspecifies the integration algorithm, where leapfrog specifies the default leap frog integration, and leapfrog_bc specifies the Brown-Clarke leap frog integrator.
[finish | resume]if specified, directs to finish or extend the simulation for which the restart file is read.
guidedspecifies the use of the guided molecular dynamics simulation technique. The current implementation is still under development.
equil <integer mequi>specifies the number of equilibration steps
<mequi>, with a default
of 100.
data <integer mdacq> [over <integer ldacq>]]specifies the number of data gathering steps
<mdacq> with a
default of 500. In multi-configuration thermodynamic integrations
<mequi> and <mdacq> are for each of the ensembles, and
variable <ldacq> specifies the minimum number of data gathering steps
in each ensemble. In regular molecular dynamics simulations <ldacq>
is not used. The default value for <ldacq> is the value of <mdacq>.
time <real stime>specifies the initial time
<stime> of a molecular simulation in ps,
with a default of 0.0.
step <real tstep>specifies the time step
<tstep> in ps, with 0.001 as the default value.
isotherm [<real tmpext>] [trelax <real tmprlx> [<real tmsrlx>]]specifies a constant temperature ensemble using Berendsen's thermostat, where
<tmpext> is the external temperature with a default of 298.15 K,
and <tmprlx> and <tmsrlx> are temperature relaxation times in ps
with a default of 0.1. If only <tmprlx> is given the complete system
is coupled to the heat bath with relaxation time <tmprlx>. If both
relaxation times are supplied, solvent and solute are independently coupled
to the heat bath with relaxation times <tmprlx> and <tmsrlx>,
respectively.
isobar [<real prsext>] [trelax <real prsrlx> ] \
[compress <real compr>] [anisotropic] [noz]
specifies a constant pressure ensemble using Berendsen's piston,
where <prsext> is the external pressure with a default of 1.025 10<prsrlx> is the pressure relaxation time in ps with a default of 0.5, and
<compr> is the system compressibility in m
vreass <integer nfgaus> <real tgauss>specifies that velocities will be reassigned every
<nfgaus> molecular
dynamics steps, reflecting a temperature of <tgauss> K. The default
is not to reassign velocities, i.e. <nfgaus> is 0.
cutoff [short] <real rshort> [long <real rlong>] \
[qmmm <real rqmmm>]
specifies the short range cutoff radius <rshort>, and the long range
cutoff radius <rlong> in nm. If the long range cutoff radius
is larger than the short range cutoff radius the twin range method will
be used, in which short range forces and energies are evaluated every
molecular dynamics step, and long range forces and energies with a
frequency of <nflong> molecular dynamics steps. Keyword
qmmm specifies the radius of the zone around quantum atoms
defining the QM/MM bare charges.
The default value for <rshort>, <rlong> and <rqmmm>
is 0.9 nm.
polar (first | scf [[<integer mpolit>] <real ptol>])specifies the use of polarization potentials, where the keyword first specifies the first order polarization model, and scf specifies the self consistent polarization field model, iteratively determined with a maximum of
<mpolit>
iterations to within a tolerance of <ptol> D in the generated
induced dipoles. The default is not to use polarization models.
field <real xfield> [freq <real xffreq>] [vector <real xfvect(1:3)>]specifies an external electrostatic field, where
<xfield> is the field strength, <xffreq> is the
frequency in MHz and <xfvect> is the external field vector.
shake [<integer mshitw> [<integer mshits>]] \
[<real tlwsha> [<real tlssha>]]
specifies the use of SHAKE constraints,
where <mshitw> is the maximum number of solvent SHAKE iterations,
and <mshits> is the maximum number of solute SHAKE iterations. If
only <mshitw> is specified, the value will also be used for <mshits>.
The default maximum number of iterations is 100 for both.
<tlwsha> is the solvent SHAKE tolerance in nm, and <tlssha> is
the solute SHAKE tolerance in nm. If only <tlwsha> is specified, the
value given will also be used for <tlssha>. The default tolerance
is 0.001 nm for both.
noshake (solvent | solute)disables SHAKE and treats the bonded interaction according to the force field.
pme [grid <integer ng>] [alpha <real ealpha>] \
[order <integer morder>] [fft <integer imfft>]
specifies the use of smooth particle-mesh Ewald long range
interaction treatment,
where ng is the number of grid points per dimension,
ealpha is the Ewald coefficient in nmmorder is order of the Cardinal B-spline
interpolation which must be an even number and at least 4 (default
value). A platform specific 3D fast Fourier transform is used, if
available, when imfft is set to 2.
( fix | free ) (solvent [<integer idfirst> [<integer idlast>]] | \
solute [<integer idfirst> [<integer idlast>]] [ heavy | {<string atomname>}] | \
permanent )
For solvent the molecule numbers idfirst and idlastmay be
specified to be the first and last molecule to which the directive
applies. If omitted, the directive applies to all molecules. For solute,
the segment numbers idfirst and idlastmay be
specified to be the first and last segment to which the directive
applies. If omitted, the directive applies to all segments. In addition,
the keyword heavy may be specified to apply to all non hydrogen
atoms in the solute, or a set of atom names may be specified in which
a wildcard character ? may be used.
auto <integer lacf> [fit <integer nfit>] [weight <real weight>]controls the calculation of the autocorrelation, where
<lacf> is the length of the autocorrelation function, with
a default of 1000, <nfit> is the number of functions used in the
fit of the autocorrelation function, with a default of 15, and
<weight> is the weight factor for the autocorrelation function,
with a default value of 0.0.
print topol [nonbond] [solvent] [solute]specifies printing the topology information, where nonbond refers to the non-bonded interaction parameters, solvent to the solvent bonded parameters, and solute to the solute bonded parameters. If only topol is specified, all topology information will be printed to the output file.
print step <integer nfoutp> [extra] [energy]specifies the frequency
nfoutp of printing molecular dynamics step
information to the output file. If the keyword extra is specified
additional energetic data are printed for solvent and solute separately.
If the keyword energy is specified, information is printed for
all bonded solute interactions.
The default for nfoutp is 0. For molecular dynamics simulations
this frequency is in time steps, and for multi-configuration thermodynamic
integration in
print stat <integer nfstat>specifies the frequency
<nfstat> of printing statistical information
of properties that are calculated during the simulation.
For molecular dynamics simulation
this frequency is in time steps, for multi-configuration thermodynamic
integration in
update pairs <integer nfpair>specifies the frequency
<nfpair> in molecular dynamics steps of
updating the pair lists. The default for the frequency is 1.
In addition, pair lists are also updated after each step in which
recording of the restart or trajectory files is performed. Updating
the pair lists includes the redistribution of atoms that changed
domain and load balancing, if specified.
update long <integer nflong>specifies the frequency
<nflong> in molecular dynamics steps
of updating the long range forces. The default frequency is 1.
The distinction of short range and long range forces is only
made if the long range cutoff radius was specified to be larger
than the short range cutoff radius. Updating the long range forces
is also done in every molecular dynamics step in which the
pair lists are regenerated.
update center <integer nfcntr> [zonly] [fraction <integer idscb(1:5)>]specifies the frequency
<nfcntr> in molecular dynamics steps in
which the center of geometry of the solute(s) is translated to the
center of the simulation volume. Optional keyword zonly can be
used to specify that centering will take place in the z-direction
only.
The solute fractions determining the
solutes that will be centered are specified by the keyword
fraction and the vector <idscb>, with a maximum of 5 entries.
This translation is implemented such that it has no effect on any
aspect of the simulation. The default is not to center, i.e. nfcntr is
0. The default fraction used to center solute is 1.
update motion <integer nfslow>specifies the frequency
<nfslow> in molecular dynamics steps of
removing the center of mass motion.
update analysis <integer nfanal>specifies the frequency
<nfanal> in molecular dynamics steps of
invoking the analysis module.
update rdf <integer nfrdf> [range <real rrdf>] [bins <integer ngl>]specifies the frequency
<nfrdf> in molecular dynamics steps of
calculating contributions to the radial distribution functions.
The default is 0. The range of the radial distribution
functions is given by <rrdf> in nm, with a default of the short
range cutoff radius. Note that radial distribution functions are not
evaluated beyond the short range cutoff radius. The number of
bins in each radial distribution function is given by <ngl>, with
a default of 1000.
If radial distribution function are to be
calculated, a rdi files needs to be available in which the
contributions are specified as follows.
| Card | Format | Description |
| I-1 | i | Type, 1=solvent-solvent, 2=solvent-solute, 3-solute-solute |
| I-2 | i | Number of the rdf for this contribution |
| I-3 | i | First atom number |
| I-4 | i | Second atom number |
record rest <integer nfrest> [keep]specifies the frequency
<nfrest> in molecular dynamics steps
of rewriting the restart file, with extension rst.
For multi-configuration
thermodynamic integration simulations the frequency is in
steps in
record coord <integer nfcoor>specifies the frequency
<nfcoor> in molecular dynamics steps
of writing coordinates to the trajectory file. This directive redefines
previous record coord, record wcoor and record scoor
directives.
The default is not to record.
record wcoor <integer nfwcoo>specifies the frequency
<nfcoor> in molecular dynamics steps
of writing solvent coordinates to the trajectory file. This keyword
takes precedent over record coord. This directive redefines
previous record coord, record wcoor and record scoor
directives.
The default is not to record.
record scoor <integer nfscoo>specifies the frequency
<nfscoo> in molecular dynamics steps
of writing solute coordinates to the trajectory file. This keyword
takes precedent over record coord. This directive redefines
previous record coord, record wcoor and record scoor
directives.
The default is not to record.
record veloc <integer nfvelo>specifies the frequency
<nfvelo> in molecular dynamics steps
of writing velocitiesto the trajectory file. This directive redefines
previous record veloc, record wvelo and record svelo
directives.
The default is not to record.
record wvelo <integer nfwvel>specifies the frequency
<nfvelo> in molecular dynamics steps
of writing solvent velocitiesto the trajectory file. This keyword
takes precedent over record veloc. This directive redefines
previous record veloc, record wvelo and record svelo
directives.
The default is not to record.
record svelo <integer nfsvel>specifies the frequency
<nfsvel> in molecular dynamics steps
of writing solute velocities to the trajectory file. This keyword
takes precedent over record veloc. This directive redefines
previous record veloc, record wvelo and record svelo
directives.
The default is not to record.
record prop <integer nfprop>specifies the frequency
<nfprop> in molecular dynamics steps
of writing information to the property file, with extension
prp. For multi-configuration
thermodynamic integration simulations the frequency is in
steps in
record free <integer nffree>specifies the frequency
<nffree> in multi-configuration
thermodynamic integration steps to record data to the
free energy data file, with extension gib.
The default is 1, i.e. to record at every
record sync <integer nfsync>specifies the frequency
<nfsync> in molecular dynamics steps
of writing information to the synchronization file, with extension
syn.
The default is not to record.
The information written is the simulation time, the wall clock time
of the previous MD step, the wall clock time of the previous force
evaluation, the total synchronization time, the largest
synchronization time and the node on which the largest synchronization
time was found. The recording of synchronization times is part of the
load balancing algorithm. Since load balancing is only performed when
pair-lists are updated, the frequency <nfsync> is correlated
with the frequency of pair-list updates <nfpair>. This directive
is only needed for analysis of the load balancing performance. For
normal use this directive is not used.
record times <integer nftime>specifies the frequency
<nfsync> in molecular dynamics steps
of writing information to the timings file, with extension
tim.
The default is not to record.
The information written is wall clock time used by each of the
processors for the different components in the force evaluation.
This directive is only needed for analysis of the wall clock time
distribution. For normal use this directive is not used.
load [reset] ( none | size [<real factld>] | pairs |
(pairs [<integer ldpair>] size [<real factld>]) )
determines the type of dynamic load balancing performed,
where the default is none. Load balancing option size
is resizing boxes on a node, and pairs redistributes the
box-box interactions over nodes. Keyword reset will reset the
load balancing read from the restart file. The level of box resizing
can be influenced with 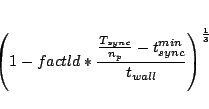 |
(27.1) |
ldpair is the number of successive pair
redistribution load balancing steps in which the accumulated synchronization
time increases, before a resizing load balancing step will be attempted.
Load balancing is only performed in molecular dynamics steps in which the
pair-list is updated. The default is not to use load balancing. In most
cases, effective load balancing is obtained with
load pairs 10 size 0.75
nodes <integer npx> <integer npy> <integer npz>specifies the distribution of the available nodes over the three Cartesian dimensions. The default distribution is chosen such that,
<npx><npy><npz>=<np>
and <npx> <npy> <npz>,
where <npx>, <npy> and <npz> are the nodes in the
x, y and z dimension respectively, and <np> is the number of nodes
allocated for the calculation. Where more than one combination
of <npx>, <npy> and <npz> are possible, the
combination is chosen with the minimum value of
<npx><npy><npz>. To change the default setting
the following optional input option is provided.
boxes <integer nbx> <integer nby> <integer nbz>specifies the distribution of boxes, where
<nbx>, <nby> and <nbz> are the number of
boxes in x, y and z direction, respectively.
The molecular system is decomposed into boxes that form the smallest
unit for communication of atomic data between nodes. The size of the
boxes is per default set to the short-range cutoff radius. If
long-range cutoff radii are used the box size is set to half the
long-range cutoff radius if it is larger than the short-range cutoff.
If the number of boxes in a dimension is less than the number of
processors in that dimension, the number of boxes is set to the number
of processors.
extra <integer madbox>sets the number of additional boxes for which memory is allocated. In rare events the amount of memory set aside per node is insufficient to hold all atomic coordinates assigned to that node. This leads to execution which aborts with the message that mwm or msa is too small. Jobs may be restarted with additional space allocated by where
<madbox> is the number of additional boxes that are allocated
on each node. The default for <madbox> is 6.
In some cases <madbox> can be reduced to 4 if memory usage is a
concern. Values of 2 or less will almost certainly result in memory
shortage.
mwm <integer mwmreq>sets the maximum number of solvent molecules
<mwmreq> per node,
allowing increased memory to be allocated for solvent molecules. This
option can be used if execution aborted because mwm was too
small.
msa <integer msareq>sets the maximum number of solute atoms
<msareq> per node,
allowing increased memory to be allocated for solute atoms. This
option can be used if execution aborted because msa was too
small.
mbb <integer mbbreq>sets the maximum number of sub-box pairs
<mbbreq> per node,
allowing increased memory to be allocated for the sub-box pair lists.
This option can be used if execution aborted because mbbl was too
small.
memory <integer memlim>sets a limit
<memlim> in kB on the allocated amount of memory used by
the molecular dynamics module.
Per default all available memory is allocated. Use of this command
is required for QM/MM simulations only.