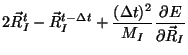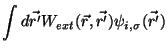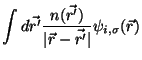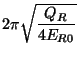


Next: 36. Controlling NWChem with
Up: user
Previous: 34. File formats
Contents
Subsections
The NWChem plane-wave (NWPW) module uses pseudopotentials and
plane-wave basis sets to perform Density Functional Theory
calculations. This module complements the capabilities of the more
traditional Gaussian function based approaches (i.e. DFT module and
GAPPS Module) by having an accuracy at least as good as the
traditional Gaussian function based approaches for many applications,
yet is still fast enough to treat systems containing hundreds of
atoms. Another significant advantage is its ability to simulate
dynamics on a ground state potential surface directly at run-time
using the Car-Parrinello algorithm. This method's efficiency and
accuracy make it a desirable first principles method of simulation in
the study of complex molecular, liquid, and solid state systems.
Applications for this first principles method include the calculation
of free energies, search for global minima, explicit simulation of
solvated molecules, and simulations of complex vibrational modes that
cannot be described within the harmonic approximation.
The NWPW module is a collection of three modules.
- PSPW - (PSeudopotential Plane-Wave) A gamma point code for
calculating molecules, liquids, crystals, and surfaces.
- Band - A band structure code for calculating
crystals and surfaces with small band gaps (e.g. semi-conductors
and metals).
- PAW - a prototype (gamma point) projector augmented plane-wave code
for calculating molecules, crystals, and surfaces
The PSPW, Band, and PAW modules can be used to compute the energy and optimize the
geometry. Both the PSPW and Band modules can also be used to find saddle points, and
compute numerical second derivatives. In addition the PSPW module can also be used
to perform Car-Parrinello molecular dynamics.
Section 35.1 describes the tasks contained within the
PSPW module, section 35.2 describes the tasks
contained within the Band module, section 35.3 describes
the tasks contained within the PAW module, and section 35.4
describes the pseudopotential library included with NWChem. The
datafiles used by the PSPW module are described in section
35.5. Car-Parrinello output data files are described
in section 35.5.8, and the minimization and
Car-Parrinello algorithms are described in sections
![[*]](crossref.png) -35.6. Examples of how
to setup and run a PSPW geometry optimization, a Car-Parrinello
simulation, a band structure minimization, and a PAW geometry
optimization are presented in sections 35.7, 35.8, and
35.11, and 35.12.
Finally in section 35.13 the capabilities and limitations of the NWPW module are discussed.
-35.6. Examples of how
to setup and run a PSPW geometry optimization, a Car-Parrinello
simulation, a band structure minimization, and a PAW geometry
optimization are presented in sections 35.7, 35.8, and
35.11, and 35.12.
Finally in section 35.13 the capabilities and limitations of the NWPW module are discussed.
If you are a first time user of this module it is recommended that you skip the next five sections and proceed directly to the tutorials in sections
35.7-35.12.
35.1 PSPW Tasks
All input to the PSPW Tasks is contained within the compound PSPW block,
PSPW
...
END
To perform an actual calculation a TASK PSPW directive is used
(Section 5.10).
TASK PSPW
In addition to the directives listed in Section 5.10, i.e.
TASK pspw energy
TASK pspw gradient
TASK pspw optimize
TASK pspw saddle
TASK pspw freqencies
TASK pspw vib
there are additional directives that are specific to the PSPW module, which are:
TASK PSPW [Car-Parrinello ||
pspw_dplot ||
wannier ||
psp_generator ||
steepest_descent ||
psp_formatter ||
wavefunction_initializer ||
v_wavefunction_initializer ||
wavefunction_expander ]
Once a user has specified a geometry, the PSPW module can be invoked
with no input directives (defaults invoked throughout). However, the
user will probably always specify the simulation cell used in the
computation, since the default simulation cell is not well suited for
most systems. There are sub-directives which allow for customized
application; those currently provided as options for the PSPW module are:
PSPW
CELL_NAME <string cell_name default 'cell_default'>
INPUT_WAVEFUNCTION_FILENAME <string input_wavefunctions default input_movecs>
OUTPUT_WAVEFUNCTION_FILENAME <string output_wavefunctions default input_movecs>
FAKE_MASS <real fake_mass default 400000.0>
TIME_STEP <real time_step default 5.8>
LOOP <integer inner_iteration outer_iteration default 10 100>
TOLERANCES <real tole tolc default 1.0e-7 1.0e-7>
ENERGY_CUTOFF <real ecut default (see input description)>
WAVEFUNCTION_CUTOFF <real wcut default (see input description)>
EWALD_NCUT <integer ncut default 1>]
EWALD_RCUT <real rcut default (see input description)>
XC (Vosko || PBE96 default Vosko)
DFT||ODFT||RESTRICTED||UNRESTRICTED
MULT <integer mult default 1>
MULLIKEN
ALLOW_TRANSLATION
SIMULATION_CELL ... (see input description) END
DPLOT ... (see input description) END
WANNIER ... (see input description) END
CAR-PARRINELLO ... (see input description) END
PSP_GENERATOR ... (see input description) END
WAVEFUNCTION_INITIALIZER ... (see input description) END
V_WAVEFUNCTION_INITIATIZER ... (see input description) END
WAVEFUNCTION_EXPANDER ... (see input description) END
STEEPEST_DESCENT ... (see input description) END
END
The following list describes the keywords contained in the PSPW input block.
A prototype limited memory BFGS (LMBFGS) minimizer can be used to minimize the energy. To
use this new optimizer the following SET directive needs to be specified:
set nwpw:mimimizer 1 # Default - Grassman conjugate gradient minimizer is used to minimize the energy.
set nwpw:mimimizer 2 # Grassman LMBFGS minimimzer is used to minimize the energy.
set nwpw:minimizer 4 # Stiefel conjugate gradient minimizer is used to minimize the energy.
set nwpw:minimizer 5 # Band-by-band minimizer is used to minimize the energy.
Limited testing suggests that the Grassman LMBFGS minimizer is about twice as fast as
the conjugate gradient minimizer. However, there are several known cases
where this optimizer fails, so it is currently not a default option, and
should be used with caution.
In addition the following SET directives can be specified:
set nwpw:lcao_skip .false. # Default - initial wavefunctions generated using an LCAO guess.
set nwpw:lcao_skip .true. # Initial wavefunctions generated using a random plane-wave guess.
set nwpw:lcao_print .false. # Default - Ouput not produced during the generation of the LCAO guess.
set nwpw:lcao_print .true. # Output produced during the generation of the LCAO guess.
set nwpw:lcao_iterations 2 #specifies the number of LCAO iterations
35.1.1 Simulation Cell
Simulation cells are stored in the RTDB. To enter a simulation cell
into the RTDB the user defines a simulation_cell sub-block within the PSPW
block. Listed below is the format of a simulation_cell sub-block.
PSPW
...
SIMULATION_CELL
CELL_NAME <string name default 'cell_default'>
BOUNDARY_CONDITIONS (periodic || aperiodic default periodic)
LATTICE_VECTORS
<real a1.x a1.y a1.z default 20.0 0.0 0.0>
<real a2.x a2.y a2.z default 0.0 20.0 0.0>
<real a3.x a3.y a3.z default 0.0 0.0 20.0>
NGRID <integer na1 na2 na3 default 32 32 32>
END
...
END
Basically, the user needs to enter the dimensions, gridding and boundry
conditions of the simulation cell. The following list describes the
input in detail.
 name
name - user-supplied name for the simulation block.
- user-supplied name for the simulation block.
- periodic - keyword specifying that the simulation cell
has periodic boundary conditions.
- aperiodic - keyword specifying that the simulation cell
has free-space boundary conditions.
 a1.x a1.y a1.z
a1.x a1.y a1.z - user-supplied values for the first
lattice vector
- user-supplied values for the first
lattice vector
 a2.x a2.y a2.z
a2.x a2.y a2.z - user-supplied values for the second
lattice vector
- user-supplied values for the second
lattice vector
 a3.x a3.y a3.z
a3.x a3.y a3.z - user-supplied values for the third
lattice vector
- user-supplied values for the third
lattice vector
 na1 na2 na3
na1 na2 na3 - user-supplied values for discretization
along lattice vector directions.
- user-supplied values for discretization
along lattice vector directions.
Unit Cell Optimization
The PSPW module using the DRIVER geometry optimizer can optimize a crystal unit cell.
Currently this type of optimization works only if the geometry is specified in fractional
coordinates. The following SET directive is used to tell the DRIVER geometry optimizer to
optimize the crystal unit cell in addition to the geometry.
set includestress .true.
35.1.3 DPLOT
The pspw dplot task is used to generate plots of various types of electron
densities (or orbitals) of a molecule. The electron density is calculated on the
specified set of grid points from a PSPW calculation. The output file
generated is in the Gaussian Cube format.
Input to the DPLOT task is contained
within the DPLOT sub-block.
PSPW
...
DPLOT
...
END
...
END
To run a DPLOT calculation the following directive
is used:
TASK PSPW PSPW_DPLOT
Listed below is the format of a DPLOT sub-block.
PSPW
...
DPLOT
VECTORS <string input_wavefunctions default input_movecs>
DENSITY [total||difference||alpha||beta||laplacian||potential default total] <string density_name no default>
ELF [restricted|alpha|beta] <string elf_name no default>
ORBITAL <integer orbital_number no default> <string orbital_name no default>
END
...
END
The following list describes the input for the DPLOT
sub-block.
 input_wavefunctions
input_wavefunctions - name of pspw wavefunction file.
- name of pspw wavefunction file.
 density_name
density_name - filename containing Gaussian Cube file of
density plot.
- filename containing Gaussian Cube file of
density plot.
 elf_name
elf_name - filename containing Gaussian Cube file of and
ELF plot
- filename containing Gaussian Cube file of and
ELF plot
 orbital_name
orbital_name - filename containing Gaussian Cube file of
orbital plot.
- filename containing Gaussian Cube file of
orbital plot.
35.1.4 Wannier
The pspw wannier task is generate maximally localized (Wannier) molecular orbitals. The
algorithm proposed by Silvestrelli et al is use to generate the Wannier orbitals. The
current version of this code works only for cubic cells.
Input to the Wannier task is contained within the Wannier sub-block.
PSPW
...
Wannier
...
END
...
END
To run a Wannier calculation the following directive
is used:
TASK PSPW Wannier
Listed below is the format of a Wannier sub-block.
PSPW
...
Wannier
OLD_WAVEFUNCTION_FILENAME <string input_wavefunctions default input_movecs>
NEW_WAVEFUNCTION_FILENAME <string output_wavefunctions default input_movecs>
END
...
END
The following list describes the input for the Wannier
sub-block.
 input_wavefunctions
input_wavefunctions - name of pspw wavefunction file.
- name of pspw wavefunction file.
 output_wavefunctions
output_wavefunctions - name of pspw wavefunction file that
will contain the Wannier orbitals.
- name of pspw wavefunction file that
will contain the Wannier orbitals.
35.1.5 Self-Interaction Corrections
The SET directive is used to specify the molecular orbitals
contribute to the self-interaction-correction (SIC) term.
set pspw:SIC_orbitals <integer list_of_molecular_orbital_numbers>
This defines only the molecular orbitals in the list as SIC active. All
other molecular orbitals will not contribute to the SIC term.
For example the following directive specifies that the molecular orbitals numbered
1,5,6,7,8, and 15 are SIC active.
set pspw:SIC_orbitals 1 5:8 15
or equivalently
set pspw:SIC_orbitals 1 5 6 7 8 15
The following directive turns on self-consistent SIC.
set pspw:SIC_relax .false. # Default - Perturbative SIC calculation
set pspw:SIC_relax .true. # Self-consistent SIC calculation
Two types of solvers can be used and they are specified using the following
SET directive
set pspw:SIC_solver_type 1 # Default - cutoff coulomb kernel
set pspw:SIC_solver_type 2 # Free-space boundary condition kernel
The parameters for the cutoff coulomb kernel are defined by the following
SET directives:
set pspw:SIC_screening_radius <real rcut>
set pspw:SIC_screening_power <real rpower>
35.1.6 Point Charge Analysis
The MULLIKEN option can be used to generate derived atomic point charges
from a plane-wave density. This analysis is based on a strategy suggested in the work of
P.E. Blochl, J. Chem. Phys. vol. 103, page 7422 (1995). In this strategy
the low-frequency components a plane-wave density are fit to a linear
combination of atom centered Gaussian functions.
The following SET directives are used to define the fitting.
set pspw_APC:Gc <real Gc_cutoff> # specifies the maximum frequency component of the density to be used in the fitting in units of au.
set pspw_APC:nga <integer number_gauss> # specifies the the number of Gaussian functions per
atom.
set pspw_APC:gamma <real gamma_list> # specifies the decay lengths of each atom centered Gaussian.
We suggest using the following parameters.
set pspw_APC:Gc 2.5
set pspw_APC:nga 3
set pspw_APC:gamma 0.6 0.9 1.35
35.1.7 Car-Parrinello
The Car-Parrinello task is used to perform ab initio molecular dynamics
using the scheme developed by Car and Parrinello. In this unified ab
initio molecular dynamics scheme the motion of the ion cores is coupled to
a fictitious motion for the Kohn-Sham orbitals of density functional
theory. Constant energy or constant temperature simulations can be
performed. A detailed description of this method
is described in section 35.6.
Input to the Car-Parrinello simulation is contained within the
Car-Parrinello sub-block.
PSPW
...
Car-Parrinello
...
END
...
END
To run a Car-Parrinello calculation the following directive is used:
TASK PSPW Car-Parrinello
The Car-Parrinello sub-block contains a great deal
of input, including pointers to data, as well as
parameter input. Listed below is the format of a Car-Parrinello sub-block.
PSPW
...
Car-Parrinello
CELL_NAME <string cell_name default 'cell_default'>
INPUT_WAVEFUNCTION_FILENAME <string input_wavefunctions default input_movecs>
OUTPUT_WAVEFUNCTION_FILENAME <string output_wavefunctions default input_movecs>
INPUT_V_WAVEFUNCTION_FILENAME <string input_v_wavefunctions default input_vmovecs>
OUTPUT_V_WAVEFUNCTION_FILENAME <string output_v_wavefunctions default input_vmovecs>
FAKE_MASS <real fake_mass default default 1000.0>
TIME_STEP <real time_step default 5.0>
LOOP <integer inner_iteration outer_iteration default 10 1>
SCALING <real scale_c scale_r default 1.0 1.0>
ENERGY_CUTOFF <real ecut default (see input description)>
WAVEFUNCTION_CUTOFF <real wcut default (see input description)>
EWALD_NCUT <integer ncut default 1>
EWALD_RCUT <real rcut default (see input description)>
XC (Vosko || PBE96 default Vosko)
[Nose-Hoover <real Period_electron Temperature_electrion Period_ion Temperature_ion
default 100.0 298.15 100.0 298.15>]
[SA_decay <real sa_scale_c sa_scale_r default 1.0 1.0>]
XYZ_FILENAME <string xyz_filename default XYZ>
EMOTION_FILENAME <string emotion_filename default EMOTION>
HMOTION_FILENAME <string hmotion_filename default HMOTION>
OMOTION_FILENAME <string omotion_filename default OMOTION>
EIGMOTION_FILENAME <string eigmotion_filename default EIGMOTION>
ION_MOTION_FILENAME <string ion_motion_filename default MOTION>
END
...
END
The following list describes the input for the Car-Parrinello
sub-block.
When a DPLOT sub-block is specified the following SET directive can be used
to output dplot data during a Car-Parrinello simulation:
set pspw_dplot:iteration_list <integer list_of_iteration_numbers>
The Gaussian cube files specified in the DPLOT sub-block are appended
with the specified iteration number.
For example, the following directive specifies that at the
3,10,11,12,13,14,15, and 50 iterations Gaussian cube files are to be produced.
set pspw_dplot:iteration_list 3,10:15,50
35.1.8 PSP_GENERATOR
A one-dimensional pseudopotential code has been integrated into NWChem.
This code allows the user to modify and develop pseudopotentials. Currently,
only the Hamann and Troullier-Martins norm-conserving pseudopotentials can be
generated. In future releases, the pseudopotential library (section 35.4)
will be more complete, so that the user will not have explicitly generate
pseudopotentials using this module.
Input to the PSP_GENERATOR task is contained within the
PSP_GENERATOR sub-block.
PSPW
...
PSP_GENERATOR
...
END
...
END
To run a PSP_GENERATOR calculation the following directive
is used:
TASK PSPW PSP_GENERATOR
Listed below is the format of a PSP_GENERATOR sub-block.
PSPW
...
PSP_GENERATOR
PSEUDOPOTENTIAL_FILENAME: <string psp_name>
ELEMENT: <string element>
CHARGE: <real charge>
MASS_NUMBER: <real mass_number>
ATOMIC_FILLING: <integer ncore nvalence>
( (1||2||...) (s||p||d||f||...) <real filling> \
...)
[CUTOFF: <integer lmax>
( (s||p||d||f||g) <real rcut>\
...)
]
PSEUDOPOTENTIAL_TYPE: (TROULLIER-MARTINS || HAMANN default HAMANN)
SOLVER_TYPE: (PAULI || SCRHODINGER default PAULI)
EXCHANGE_TYPE: (dirac || PBE96 default DIRAC)
CORRELATION_TYPE: (VOSKO || PBE96 default VOSKO)
[SEMICORE_RADIUS: <real rcore>]
end
...
END
The following list describes the input for the PSP_GENERATOR
sub-block.
 psp_name
psp_name - name that points to a.
- name that points to a.
 element
element - Atomic symbol.
- Atomic symbol.
 charge
charge - charge of the atom
- charge of the atom
 mass
mass - mass number for the atom
- mass number for the atom
 ncore
ncore - number of core states
- number of core states
 nvalence
nvalence - number of valence states.
- number of valence states.
- ATOMIC_FILLING:.....(see below)
 filling
filling - occupation of atomic state
- occupation of atomic state
- CUTOFF:....(see below)
 rcore
rcore - value for the semicore radius (see below)
- value for the semicore radius (see below)
This required block is used to define the reference atom which is used
to define the pseudopotential. After the ATOMIC_FILLING:  ncore
ncore
 nvalence
nvalence line, the core states are listed (one per line), and
then the valence states are listed (one per line).
Each state contains two integer and a value. The first integer
specifies the radial quantum number,
line, the core states are listed (one per line), and
then the valence states are listed (one per line).
Each state contains two integer and a value. The first integer
specifies the radial quantum number, 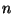 ,
The second integer specifies the angular momentum quantum number,
,
The second integer specifies the angular momentum quantum number, 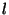 ,
and the third value specifies the occupation of the state.
,
and the third value specifies the occupation of the state.
For example to define a pseudopotential
for the Neon atom in the
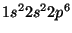 state
could have the block
state
could have the block
ATOMIC_FILLING: 1 2
1 s 2.0 #core state - 1s^2
2 s 2.0 #valence state - 2s^2
2 p 6.0 #valence state - 2p^6
for a pseudopotential with a 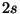 and
and 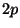 valence electrons
or the block
valence electrons
or the block
ATOMIC_FILLING: 3 0
1 s 2.0 #core state
2 s 2.0 #core state
2 p 6.0 #core state
could be used for a pseudopotential with no valence electrons.
This optional block specifies the cutoff distances used
to match the all-electron atom to the pseudopotential atom. For
Hamann pseudopotentials 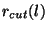 defines the distance
where the all-electron potential is matched to the pseudopotential, and
for Troullier-Martins pseudopotentials
defines the distance
where the all-electron potential is matched to the pseudopotential, and
for Troullier-Martins pseudopotentials 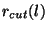 defines the distance
where the all-electron orbital is matched to the pseudowavefunctions.
Thus the definition of the radii depends on the type of pseudopotential.
The cutoff radii used in Hamann pseudopotentials will be smaller than
the cufoff raddi used in Troullier-Martins pseudopotentials.
defines the distance
where the all-electron orbital is matched to the pseudowavefunctions.
Thus the definition of the radii depends on the type of pseudopotential.
The cutoff radii used in Hamann pseudopotentials will be smaller than
the cufoff raddi used in Troullier-Martins pseudopotentials.
For example to define a softened Hamann pseudopotential for
Carbon would be
ATOMIC_FILLING: 1 2
1 s 2.0
2 s 2.0
2 p 2.0
CUTOFF: 2
s 0.8
p 0.85
d 0.85
while a similarly softened Troullier-Marting pseudopotential
for Carbon would be
ATOMIC_FILLING: 1 2
1 s 2.0
2 s 2.0
2 p 2.0
CUTOFF: 2
s 1.200
p 1.275
d 1.275
Specifying the SEMICORE_RADIUS option turns on the semicore correction approximation proposed
by Louie et al (S.G. Louie, S. Froyen, and M.L. Cohen, Phys. Rev. B, 26, 1738, (1982)).
This approximation is known to dramatically improve results for systems containing
alkali and transition metal atoms.
The implmentation in the PSPW module defines the semi-core denisty,
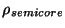 in terms of
the core density,
in terms of
the core density, 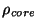 , by using the sixth-order polynomial
, by using the sixth-order polynomial
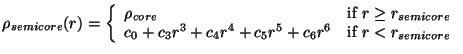 |
|
|
(35.1) |
This expansion was suggested by Fuchs and Scheffler
(M. Fuchs, and M. Scheffler, Comp. Phys. Comm.,119,67 (1999)),
and is better behaved for taking derivatives (i.e. calculating ionic forces) than the expansion suggested
by Louie et al.
35.1.9 WAVEFUNCTION_INTITIALIZER
The functionality of this task is now performed automatically. For backwards
compatibility, we provide a description of the input to this task.
The wavefunction_initializer task is used to generate an initial wavefunction
datafile.
Input to the WAVEFUNCTION_INITIALIZER task is contained
within the WAVEFUNCTION_INITIALIZER sub-block.
PSPW
...
WAVEFUNCTION_INITIALIZER
...
END
...
END
To run a WAVEFUNCTION_INITIALIZER calculation the following directive
is used:
TASK PSPW WAVEFUNCTION_INITIALIZER
Listed below is the format of a WAVEFUNCTION_INITIALIZER sub-block.
PSPW
...
WAVEFUNCTION_INITIALIZER
CELL_NAME: <string cell_name>
WAVEFUNCTION_FILENAME: <string wavefunction_name default input_movecs>
(RESTRICTED||UNRESTRICTED)
if (RESTRICTED)
RESTRICTED_ELECTRONS: <integer restricted electrons>
if (UNRESTRICTED)
UP_ELECTRONS: <integer up_electrons>
DOWN_ELECTRONS: <integer down_electrons>
END
...
END
The following list describes the input for the WAVEFUNCTION_INITIALIZER
sub-block.
For backwards compatibility, the input to the WAVEFUNCTION_INITIALIZER
sub-block can also be of the form
PSPW
...
WAVEFUNCTION_INITIALIZER
CELL_NAME: <string cell_name>
WAVEFUNCTION_FILENAME: <string wavefunction_name default input_movecs>
(RESTRICTED||UNRESTRICTED)
[UP_FILLING: <integer up_filling>
[0 0 0 0]
{<integer kx ky kz> (-2||-1||1||2)}]
[DOWN_FILLING: <integer down_filling>
[0 0 0 0]
{<integer kx ky kz> (-2||-1||1||2)}]
END
...
END
where
 cell_name
cell_name - name that points
to the simulation_cell named
- name that points
to the simulation_cell named  cell_name
cell_name . See section 35.1.1.
. See section 35.1.1.
 wavefunction_name
wavefunction_name - name that will point
to a wavefunction file.
- name that will point
to a wavefunction file.
- RESTRICTED - keyword specifying that the calculation is restricted.
- UNRESTRICTED - keyword specifying that the calculation is unrestricted.
 up_filling
up_filling - number of restricted molecular orbitals if
RESTRICTED and number of spin-up molecular orbitals if
UNRESTRICTED.
- number of restricted molecular orbitals if
RESTRICTED and number of spin-up molecular orbitals if
UNRESTRICTED.
 down_filling
down_filling - number of spin-down moleclar orbitals if
UNRESTRICTED. Not used if a RESTRICTED calculation.
- number of spin-down moleclar orbitals if
UNRESTRICTED. Not used if a RESTRICTED calculation.
 kx ky kz
kx ky kz - specifies which planewave is to be filled.
- specifies which planewave is to be filled.
The values for the planewave
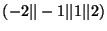 are used to represent whether
the specified planewave is a cosine or a sine function, in addition
random noise can be added to these base functions. That is
are used to represent whether
the specified planewave is a cosine or a sine function, in addition
random noise can be added to these base functions. That is 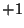 represents a cosine function, and
represents a cosine function, and 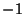 represents a sine function.
The
represents a sine function.
The 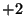 and
and 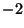 values are used to represent a cosine function with
random components added and a sine function with random components
added respectively.
values are used to represent a cosine function with
random components added and a sine function with random components
added respectively.
35.1.10 V_WAVEFUNCTION_INITIALIZER
The functionality of this task is now performed automatically. For backwards
compatibility, we provide a description of the input to this task.
The v_wavefunction_initializer task is used to generate an initial velocity
wavefunction datafile.
Input to the V_WAVEFUNCTION_INITIALIZER task is contained
within the V_WAVEFUNCTION_INITIALIZER sub-block.
PSPW
...
V_WAVEFUNCTION_INITIALIZER
...
END
...
END
To run a V_WAVEFUNCTION_INITIALIZER calculation the following directive
is used:
TASK PSPW WAVEFUNCTION_INITIALIZER
Listed below is the format of a V_WAVEFUNCTION_INITIALIZER sub-block.
PSPW
...
V_WAVEFUNCTION_INITIALIZER
V_WAVEFUNCTION_FILENAME: <string v_wavefunction_name default input_vmovecs>
CELL_NAME: <string cell_name>
(RESTRICTED||UNRESTRICTED)
UP_FILLING: <integer up_filling>
DOWN_FILLING: <integer down_filling>
END
...
END
The following list describes the input for the V_WAVEFUNCTION_INITIALIZER
sub-block.
 cell_name
cell_name - name that points
to the simulation_cell named
- name that points
to the simulation_cell named  cell_name
cell_name . See section 35.1.1.
. See section 35.1.1.
 wavefunction_name
wavefunction_name - name that will point
to a velocity wavefunction file.
- name that will point
to a velocity wavefunction file.
- RESTRICTED - keyword specifying that the calculation is restricted.
- UNRESTRICTED - keyword specifying that the calculation is unrestricted.
 up_filling
up_filling - number of restricted velocity molecular
orbitals if RESTRICTED and number of spin-up velocity molecular
orbitals if UNRESTRICTED.
- number of restricted velocity molecular
orbitals if RESTRICTED and number of spin-up velocity molecular
orbitals if UNRESTRICTED.
 down_filling
down_filling - number of spin-down velocity moleclar
orbitals if UNRESTRICTED. Not used if a RESTRICTED calculation.
- number of spin-down velocity moleclar
orbitals if UNRESTRICTED. Not used if a RESTRICTED calculation.
35.1.11 WAVEFUNCTION_EXPANDER
The wavefunction_expander task is used to convert a new wavefunction
file that spans a larger grid space from an old wavefunction file.
Input to the WAVEFUNCTION_EXPANDER task is contained
within the WAVEFUNCTION_EXPANDER sub-block.
PSPW
...
WAVEFUNCTION_EXPANDER
...
END
...
END
To run a WAVEFUNCTION_EXPANDER calculation the following directive
is used:
TASK PSPW WAVEFUNCTION_EXPANDER
Listed below is the format of a WAVEFUNCTION_EXPANDER sub-block.
PSPW
...
WAVEFUNCTION_EXPANDER
OLD_WAVEFUNCTION_FILENAME: <string old_wavefunction_name default input_movecs>
NEW_WAVEFUNCTION_FILENAME: <string new_wavefunction_name default input_movecs>
NEW_NGRID: <integer na1 na2 na3>
END
...
END
The following list describes the input for the WAVEFUNCTION_EXPANDER
sub-block.
 old_wavefunction_name
old_wavefunction_name - name that points
to a wavefunction file.
- name that points
to a wavefunction file.
 new_wavefunction_name
new_wavefunction_name - name that will
point to a wavefunction file.
- name that will
point to a wavefunction file.
 na1 na2 na3
na1 na2 na3 - number of grid points in each dimension
for the new wavefunction file.
- number of grid points in each dimension
for the new wavefunction file.
35.1.12 STEEPEST_DESCENT
The functionality of this task is now performed automatically by the PSPW minimizer.
For backwards compatibility, we provide a description of the input to this task.
The steepest_descent task is used to optimize the one-electron orbitals
with respect to the total energy. In addition it can also be used to optimize
geometries. This method is meant to be used for coarse optimization of
the one-electron orbitals. A detailed description of the this method
is described in section ![[*]](crossref.png)
Input to the steepest_descent simulation is contained
within the steepest_descent sub-block.
PSPW
...
STEEPEST_DESCENT
...
END
...
END
To run a steepest_descent calculation the following directive is used:
TASK PSPW steepest_descent
The steepest_descent sub-block contains a great deal
of input, including pointers to data, as well as
parameter input. Listed below is the format of a STEEPEST_DESCENT sub-block.
PSPW
...
STEEPEST_DESCENT
CELL_NAME <string cell_name>
[GEOMETRY_OPTIMIZE]
INPUT_WAVEFUNCTION_FILENAME <string input_wavefunctions default input_movecs>
OUTPUT_WAVEFUNCTION_FILENAME <string output_wavefunctions default input_movecs>
FAKE_MASS <real fake_mass default 400000.0>
TIME_STEP <real time_step default 5.8>
LOOP <integer inner_iteration outer_iteration default 10 1>
TOLERANCES <real tole tolc tolr default 1.0d-9 1.0d-9 1.0d-4>
ENERGY_CUTOFF <real ecut default (see input desciption)>
WAVEFUNCTION_CUTOFF <real wcut default (see input description)>
EWALD_NCUT <integer ncut default 1>
EWALD_RCUT <real rcut default (see input description)>
XC (Vosko || PBE96 default Vosko)
[MULLIKEN]
END
...
END
The following list describes the input for the STEEPEST_DESCENT
sub-block.
 cell_name
cell_name - name that points to the
the simulation_cell named
- name that points to the
the simulation_cell named  cell_name
cell_name . See section 35.1.1.
. See section 35.1.1.
- GEOMETRY_OPTIMIZE - optional keyword which if specified
turns on geometry optimization.
 input_wavefunctions
input_wavefunctions - name that points
to a file containing one-electron orbitals
- name that points
to a file containing one-electron orbitals
 output_wavefunctions
output_wavefunctions - name that will
point to file containing the one-electron orbitals at the
end of the run.
- name that will
point to file containing the one-electron orbitals at the
end of the run.
 fake_mass
fake_mass - value for the electronic
fake mass (
- value for the electronic
fake mass (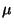 ).
).
 time_step
time_step - value for the time step (
- value for the time step (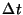 ).
).
 inner_iteration
inner_iteration - number of iterations between the
printing out of energies and tolerances
- number of iterations between the
printing out of energies and tolerances
 outer_iteration
outer_iteration - number of outer iterations
- number of outer iterations
 tole
tole - value for the energy tolerance.
- value for the energy tolerance.
 tolc
tolc - value for the one-electron orbital tolerance.
- value for the one-electron orbital tolerance.
 tolr
tolr - value for the ion position tolerance.
- value for the ion position tolerance.
 ecut
ecut - value for the cutoff energy used
to define the density. Default is set
to be the maximum value that will fit
within the simulation_cell
- value for the cutoff energy used
to define the density. Default is set
to be the maximum value that will fit
within the simulation_cell  cell_name
cell_name .
.
 wcut
wcut - value for the cutoff energy used
to define the one-electron orbitals. Default is set
to be the maximum value that will fit
within the simulation_cell
- value for the cutoff energy used
to define the one-electron orbitals. Default is set
to be the maximum value that will fit
within the simulation_cell  cell_name
cell_name .
.
 ncut
ncut - value for the number of unit cells
to sum over (in each direction) for the real space
part of the Ewald summation. Note Ewald summation
is only used if the simulation_cell is periodic.
- value for the number of unit cells
to sum over (in each direction) for the real space
part of the Ewald summation. Note Ewald summation
is only used if the simulation_cell is periodic.
 rcut
rcut - value for the cutoff radius used
in the Ewald summation. Note Ewald summation
is only used if the simulation_cell is periodic.
- value for the cutoff radius used
in the Ewald summation. Note Ewald summation
is only used if the simulation_cell is periodic.
Default set to be
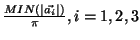 .
.
- (Vosko
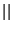 PBE96) - Choose between Vosko et al's LDA
parameterization or the Perdew, Burke,
and Erzherhoff GGA functional.
PBE96) - Choose between Vosko et al's LDA
parameterization or the Perdew, Burke,
and Erzherhoff GGA functional.
- MULLIKEN - optional keyword which if specified
causes a Mulliken anaysis to be performed at
the end of the simulation. For this option
to work angular momentum weights for each kind
of atom have to be entered into the RTDB using
the PSPW ANALYSIS sub-block (see section 35.5.3.
35.2 Band Tasks
All input to the Band Tasks is contained within the compound NWPW block,
NWPW
...
END
To perform an actual calculation a TASK Band directive is used (Section 5.10).
TASK Band
Once a user has specified a geometry, the Band module can be invoked with no input directives (defaults invoked throughout). There are sub-directives which allow for customized application; those currently provided as options for the Band module are:
NWPW
CELL_NAME <string cell_name default 'cell_default'>
ZONE_NAME <string zone_name default 'zone_default'>
INPUT_WAVEFUNCTION_FILENAME <string input_wavefunctions default input_movecs>
OUTPUT_WAVEFUNCTION_FILENAME <string output_wavefunctions default input_movecs>
FAKE_MASS <real fake_mass default 400000.0>
TIME_STEP <real time_step default 5.8>
LOOP <integer inner_iteration outer_iteration default 10 100>
TOLERANCES <real tole tolc default 1.0e-7 1.0e-7>
ENERGY_CUTOFF <real ecut default (see input description)>
WAVEFUNCTION_CUTOFF <real wcut default (see input description)>
EWALD_NCUT <integer ncut default 1>]
EWALD_RCUT <real rcut default (see input description)>
EXCHANGE_CORRELATION: (Vosko || PBE96 default Vosko)
DFT||ODFT||RESTRICTED||UNRESTRICTED
MULT <integer mult default 1>
SIMULATION_CELL ... (see input description) END
BRILLOUIN_ZONE ... (see input description) END
END
The following list describes these keywords.
 cell_name
cell_name - name that points to the
the simulation_cell named
- name that points to the
the simulation_cell named  cell_name
cell_name . See section 35.1.1.
. See section 35.1.1.
 input_wavefunctions
input_wavefunctions - name that points
to a file containing one-electron orbitals
- name that points
to a file containing one-electron orbitals
 output_wavefunctions
output_wavefunctions - name that will
point to file containing the one-electron orbitals at the
end of the run.
- name that will
point to file containing the one-electron orbitals at the
end of the run.
 fake_mass
fake_mass - value for the electronic
fake mass (
- value for the electronic
fake mass (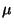 ). This parameter is not presently used in a
conjugate gradient simulation
). This parameter is not presently used in a
conjugate gradient simulation
 time_step
time_step - value for the time step (
- value for the time step (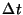 ). This
parameter is not presently used in a conjugate gradient simulation.
). This
parameter is not presently used in a conjugate gradient simulation.
 inner_iteration
inner_iteration - number of iterations between the
printing out of energies and tolerances
- number of iterations between the
printing out of energies and tolerances
 outer_iteration
outer_iteration - number of outer iterations
- number of outer iterations
 tole
tole - value for the energy tolerance.
- value for the energy tolerance.
 tolc
tolc - value for the one-electron orbital tolerance.
- value for the one-electron orbital tolerance.
 ecut
ecut - value for the cutoff energy used
to define the density. Default is set
to be the maximum value that will fit
within the simulation_cell
- value for the cutoff energy used
to define the density. Default is set
to be the maximum value that will fit
within the simulation_cell  cell_name
cell_name .
.
 wcut
wcut - value for the cutoff energy used
to define the one-electron orbitals.
Default is set to be the maximum value that
will fix within the simulation_cell
- value for the cutoff energy used
to define the one-electron orbitals.
Default is set to be the maximum value that
will fix within the simulation_cell  cell_name
cell_name .
.
 ncut
ncut - value for the number of unit cells
to sum over (in each direction) for the real space
part of the Ewald summation. Note Ewald summation
is only used if the simulation_cell is periodic.
- value for the number of unit cells
to sum over (in each direction) for the real space
part of the Ewald summation. Note Ewald summation
is only used if the simulation_cell is periodic.
 rcut
rcut - value for the cutoff radius used
in the Ewald summation. Note Ewald summation
is only used if the simulation_cell is periodic.
- value for the cutoff radius used
in the Ewald summation. Note Ewald summation
is only used if the simulation_cell is periodic.
Default set to be
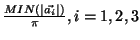 .
.
- (Vosko
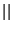 PBE96) - Choose between Vosko et al's LDA
parameterization or the Perdew, Burke,
PBE96) - Choose between Vosko et al's LDA
parameterization or the Perdew, Burke,
- SIMULATION_CELL (see section 35.1.1)
- BRILLOUIN_ZONE (see section 35.2.1)
35.2.1 Brillouin Zone
The special points of the Brillouin zone for a Band structure calculation are
stored in the RTDB. To enter special points of the Brillouin zone into the RTDB
the user defines a brillouin_zone sub-block within the NWPW
block. Listed below is the format of a brillouin_zone sub-block.
NWPW
...
BRILLOUIN_ZONE
ZONE_NAME <string name default 'zone_default'>
(KVECTOR <real k1 k2 k3 no default> <real weight default (see input description)>
...)
END
...
END
The user enters the special points and weights of the
Brillouin zone. The following list describes the input in detail.
 name
name - user-supplied name for the simulation block.
- user-supplied name for the simulation block.
 k1 k2 k3
k1 k2 k3 - user-supplied values for a special point in the
Brillouin zone.
- user-supplied values for a special point in the
Brillouin zone.
 weight
weight - user-supplied weight. Default is to set the weight
so that the sum of all the weights for the entered
special points adds up to unity.
- user-supplied weight. Default is to set the weight
so that the sum of all the weights for the entered
special points adds up to unity.
35.3 PAW Tasks
All input to the PAW Tasks is contained within the compound NWPW block,
NWPW
...
END
To perform an actual calculation the following is used (Section 5.10).
TASK PAW steepest\_descent
Once a user has specified a geometry, the PAW module can be invoked with no input directives (defaults invoked throughout). There are sub-directives which allow for customized application; those currently provided as options for the PAW module are:
NWPW
CELL_NAME <string cell_name default 'cell_default'>
[GEOMETRY_OPTIMIZE]
INPUT_WAVEFUNCTION_FILENAME <string input_wavefunctions default input_movecs>
OUTPUT_WAVEFUNCTION_FILENAME <string output_wavefunctions default input_movecs>
FAKE_MASS <real fake_mass default 400000.0>
TIME_STEP <real time_step default 5.8>
LOOP <integer inner_iteration outer_iteration default 10 100>
TOLERANCES <real tole tolc default 1.0e-7 1.0e-7>
ENERGY_CUTOFF <real ecut default (see input description)>
WAVEFUNCTION_CUTOFF <real wcut default (see input description)>
EWALD_NCUT <integer ncut default 1>]
EWALD_RCUT <real rcut default (see input description)>
EXCHANGE_CORRELATION: (Vosko || PBE96 default Vosko)
DFT||ODFT||RESTRICTED||UNRESTRICTED
MULT <integer mult default 1>
SIMULATION_CELL ... (see input description) END
END
The following list describes these keywords.
 cell_name
cell_name - name that points to the
the simulation_cell named
- name that points to the
the simulation_cell named  cell_name
cell_name . The
current version of PAW only accepts periodic unit cells.
See section 35.1.1.
. The
current version of PAW only accepts periodic unit cells.
See section 35.1.1.
- GEOMETRY_OPTIMIZE - optional keyword which if specified
turns on geometry optimization.
 input_wavefunctions
input_wavefunctions - name that points
to a file containing one-electron orbitals
- name that points
to a file containing one-electron orbitals
 output_wavefunctions
output_wavefunctions - name that will
point to file containing the one-electron orbitals at the
end of the run.
- name that will
point to file containing the one-electron orbitals at the
end of the run.
 fake_mass
fake_mass - value for the electronic
fake mass (
- value for the electronic
fake mass (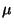 ). This parameter is not presently used in a
conjugate gradient simulation
). This parameter is not presently used in a
conjugate gradient simulation
 time_step
time_step - value for the time step (
- value for the time step (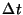 ). This
parameter is not presently used in a conjugate gradient simulation.
). This
parameter is not presently used in a conjugate gradient simulation.
 inner_iteration
inner_iteration - number of iterations between the
printing out of energies and tolerances
- number of iterations between the
printing out of energies and tolerances
 outer_iteration
outer_iteration - number of outer iterations
- number of outer iterations
 tole
tole - value for the energy tolerance.
- value for the energy tolerance.
 tolc
tolc - value for the one-electron orbital tolerance.
- value for the one-electron orbital tolerance.
 ecut
ecut - value for the cutoff energy used
to define the density. Default is set
to be the maximum value that will fit
within the simulation_cell
- value for the cutoff energy used
to define the density. Default is set
to be the maximum value that will fit
within the simulation_cell  cell_name
cell_name .
.
 wcut
wcut - value for the cutoff energy used
to define the one-electron orbitals.
Default is set to be the maximum value that
will fix within the simulation_cell
- value for the cutoff energy used
to define the one-electron orbitals.
Default is set to be the maximum value that
will fix within the simulation_cell  cell_name
cell_name .
.
 ncut
ncut - value for the number of unit cells
to sum over (in each direction) for the real space
part of the smooth compensation summation.
- value for the number of unit cells
to sum over (in each direction) for the real space
part of the smooth compensation summation.
 rcut
rcut - value for the cutoff radius used
in the smooth compensation summation.
- value for the cutoff radius used
in the smooth compensation summation.
Default set to be
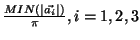 .
.
- (Vosko
 PBE96) - Choose between Vosko et al's LDA
parameterization or the Perdew, Burke,
PBE96) - Choose between Vosko et al's LDA
parameterization or the Perdew, Burke,
- SIMULATION_CELL (see section 35.1.1)
35.4 Pseudopotential and PAW basis Libraries
A library of pseudopotentials used by PSPW and BAND is currently available in the
directory
$NWCHEM_TOP/src/nwpw/libraryps/library1
The elements listed in the following table are present:
H
-------
Li Be B C N O
------- ------------------
Al Si P S
------------------------------------------------------
Zn Ga
------------------------------------------------------
Similary, a library of PAW basis used by PAW is currently available in the
directory
$NWCHEM_TOP/src/nwpw/libraryps/library2
Currently there are not very many elements available for PAW. However,
the user can request additional basis sets from Eric J. Bylaska at
(nwchem-support@emsl.pnl.gov or eric.bylaska@pnl.gov)
The elements available are: H, O Al, Sc, V, and Fe.
If you wish to redirect the code to a different directory other than
the default one,
you need to set the environmental variable
NWCHEM_NWPW_LIBRARY
to the new location of the libraryps directory.
35.5 NWPW RTDB Entries and DataFiles
Input to the PSPW and Band modules are contained in both the RTDB and datafiles.
The RTDB is used to store input that the user will need to directly specify.
Input of this kind includes ion positions, ion velocities, and simulation cell
parameters. The datafiles are used to store input, such the one-electron
orbitals, one-electron orbital velocities, formatted pseudopotentials,
and one-dimensional pseudopotentials, that the user will in most cases
run a program to generate.
The positions of the ions are stored in the default geometry structure
in the RTDB and must be specified using the GEOMETRY directive.
The velocities of the ions are stored in the default geometry structure
in the RTDB, and must be specified using the GEOMETRY directive.
35.5.3 ANALYSIS: Mulliken RTDB data
To perform Mulliken analysis information is needed from one-dimensional
pseudopotential files. In-order to facilitate the transfer of this information
to the simulation the ANALYSIS sub-block is used to exract the necessary information
and put it into the RTDB.
PSPW
...
ANALYSIS
(psp_filename: <string psp_name> \
...)
END
...
END
Basically, the user needs to enter each pseudopotential used in the simulation.
 psp_name
psp_name - name that ponts to a one-dimensional pseudopotential file.
- name that ponts to a one-dimensional pseudopotential file.
The one-electron orbitals are stored in a wavefunction datafile. This
is a binary file and cannot be directly edited. This datafile is used
by steepest_descent and Car-Parrinello tasks and can be generated
using the wavefunction_initializer or wavefunction_expander tasks.
The one-electron orbital velocities are stored in a velocity wavefunction
datafile. This is a binary file and cannot be directly edited. This datafile
is used by the Car-Parrinello task and can be generated
using the v_wavefunction_initializer task.
The pseudopotentials in Kleinman-Bylander form expanded on a simulation
cell (3d grid) are stored in a formatted pseudopotential datafile.
This is a binary file and cannot be directly edited.
This datafile
is used by steepest_descent and Car-Parrinello tasks and can be generated
using the pseudpotential_formatter task.
The one-dimensional pseudopotentials are stored in a one-dimensional
pseudopotential file. This is an ascii file and can be directly edited with
a text editor. However, the user will usually use the psp_generator
task to generate this datafile.
The data stored in the one-dimensional pseudopotential file is
character*2 element :: element name
integer charge :: valence charge of ion
real mass :: mass of ion
integer lmax :: maximum angular component
real rcut(lmax) :: cutoff radii used to define pseudopotentials
integer nr :: number of points in the radial grid
real dr :: linear spacing of the radial grid
real r(nr) :: one-dimensional radial grid
real Vpsp(nr,lmax) :: one-dimensional pseudopotentials
real psi(nr,lmax) :: one-dimensional pseudowavefunctions
real r_semicore :: semicore radius
real rho_semicore(nr) :: semicore density
and the format of it is:
[line 1: ] element
[line 2: ] charge mass lmax
[line 3: ] (rcut(l), l=1,lmax)
[line 4: ] nr dr
[line 5: ] r(1) (Vpsp(1,l), l=1,lmax)
[line 6: ] ....
[line nr+4: ] r(nr) (Vpsp(nr,l), l=1,lmax)
[line nr+5: ] r(1) (psi(1,l), l=1,lmax)
[line nr+6: ] ....
[line 2*nr+4:] r(nr) (psi(nr,l), l=1,lmax)
[line 2*nr+5:] r_semicore
if (r_semicore read) then
[line 2*nr+6:] r(1) rho_semicore(1)
[line 2*nr+7:] ....
[line 3*nr+5:] r(nr) rho_semicore(nr)
end if
35.5.8 PSPW Car-Parrinello Output Datafiles
Data file that stores ion positions and velocities as
a function of time in XYZ format.
[line 1: ] n_ion
[line 2: ]
do ii=1,n_ion
[line 2+ii: ] atom_name(ii), x(ii),y(ii),z(ii),vx(ii),vy(ii),vz(ii)
end do
[line n_ion+3 ] n_nion
do ii=1,n_ion
[line n_ion+3+ii: ] atom_name(ii), x(ii),y(ii),z(ii), vx(ii),vy(ii),vz(ii)
end do
[line 2*n_ion+4: ] ....
Datafile that stores ion positions and velocities
as a function of time
[line 1: ] it_out, n_ion, omega
[line 2: ] time
do ii=1,n_ion
[line 2+ii: ] x(ii),y(ii),z(ii), vx(ii),vy(ii),vz(ii)
end do
[line n_ion+3 ] time
do
do ii=1,n_ion
[line n_ion+3+ii: ] x(ii),y(ii),z(ii), vx(ii),vy(ii),vz(ii)
end do
[line 2*n_ion+4: ] ....
Datafile that store energies as a function of time
[line 1: ] time, E1,E2,E3,E4,E5,E6,E7,E8, (E9,E10, if Nose-Hoover)
[line 2: ] ...
Datafile that stores the rotation matrix
as a function of time.
[line 1: ] time
[line 2: ] ms,ne(ms),ne(ms)
do i=1,ne(ms)
[line 2+i: ] (hml(i,j), j=1,ne(ms)
end do
[line 3+ne(ms): ] time
[line 4+ne(ms): ] ....
Datafile that stores the eigenvalues for the one-electron
orbitals as a function of time.
[line 1: ] time, (eig(i), i=1,number_orbitals)
[line 2: ] ...
Datafile that stores a reduced representation of the
one-electron orbitals. To be used with a molecular
orbital viewer that will be ported to NWChem
in the near future.
35.6 Car-Parrinello Scheme for Ab Initio Molecular Dynamics
Car and Parrinello developed a unified scheme for doing ab initio
molecular dynamics by combining the motion of the ion cores and a ficticious
motion for the Kohn-Sham orbitals of density-functional theory
(R. Car and M. Parrinello, Phys. Rev. Lett. 55, 2471, (1985)).
At the heart of this method they introduced a ficticious kinetic energy
functional for the Kohn-Sham orbitals.
Given this kinetic energy the constrained equations of motion are found
by taking the first variation of the auxiliary Lagrangian.
Which generates a dynamics for the wavefunctions
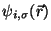 and
atoms positions
and
atoms positions 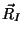 through the constrained equations of motion:
through the constrained equations of motion:
where 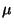 is the fictitious mass for the electronic degrees of freedom and
is the fictitious mass for the electronic degrees of freedom and
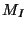 are the ionic masses.
The adjustable parameter
are the ionic masses.
The adjustable parameter 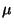 is used to
describe the relative rate at which the wavefunctions change with time.
is used to
describe the relative rate at which the wavefunctions change with time.
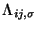 are the
Lagrangian multipliers for the orthonormalization of the single-particle
orbitals
are the
Lagrangian multipliers for the orthonormalization of the single-particle
orbitals
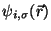 .
They are defined by the orthonormalization constraint conditions
and can be rigorously found.
However, the equations of motion for the Lagrange multipliers
depend on the specific algorithm used to integrate
Eqs. 35.4-35.5.
.
They are defined by the orthonormalization constraint conditions
and can be rigorously found.
However, the equations of motion for the Lagrange multipliers
depend on the specific algorithm used to integrate
Eqs. 35.4-35.5.
For this method to give ionic motions that are physically meaningful
the kinetic energy of the Kohn-Sham orbitals must be relatively
small when compared to the kinetic energy of the ions.
There are two ways where this criterion can fail.
First, the numerical integrations for the Car-Parrinello equations of motion
can often lead to large relative values of the kinetic energy of
the Kohn-Sham orbitals relative to the kinetic energy of the ions.
This kind of failure is easily fixed by requiring a more accurate
numerical integration, i.e. use a smaller time step for the numerical
integration.
Second, during the motion of the system a the ions can be in locations where
there is an Kohn-Sham orbital level crossing, i.e. the density-functional
energy can have two states that are nearly degenerate. This kind
of failure often occurs in the study of chemical reactions.
This kind of failure is not easily fixed and requires the use
of a more sophisticated density-functional energy that accounts
for low-lying excited electronic states.
Eqs. 35.4-35.5 integrated using the Verlet algorithm
results in
In this molecular dynamic procedure we have to know variational derivative
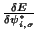 and the matrix
and the matrix
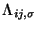 .
The variational derivative
.
The variational derivative
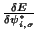 can be analytically found and is
can be analytically found and is
To find the matrix
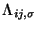 we impose the orthonormality
constraint on
we impose the orthonormality
constraint on
 to obtain a
matrix Riccatti equation, and then Riccatti equation is solved by an iterative
solution (see section
to obtain a
matrix Riccatti equation, and then Riccatti equation is solved by an iterative
solution (see section ![[*]](crossref.png) ).
).
35.6.2 Constant Temperature Simulations: Nose-Hoover Thermostats
Nose-Hoover Thermostats for the electrons and ions can also be added to the
Car-Parrinello simulation. In this type of simulation thermostats variables 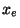 and
and 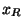 are added to the simulation by adding the auxillary energy functionals to the total energy.
are added to the simulation by adding the auxillary energy functionals to the total energy.
In these equations, the average kinetic energy for the ions is
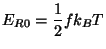 |
|
|
(35.11) |
where 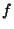 is the number of atomic degrees of freedom,
is the number of atomic degrees of freedom, 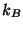 is
Boltzmans constant, and T is the desired temperature. Defining
the average ficticious kinetic energy of the electrons is not as straighforward.
Blöchl and Parrinello
(P.E. Blöchl and M. Parrinello, Phys. Rev. B, 45, 9413, (1992))
have suggested the following formula for determining
the average ficticious kinetic energy
is
Boltzmans constant, and T is the desired temperature. Defining
the average ficticious kinetic energy of the electrons is not as straighforward.
Blöchl and Parrinello
(P.E. Blöchl and M. Parrinello, Phys. Rev. B, 45, 9413, (1992))
have suggested the following formula for determining
the average ficticious kinetic energy
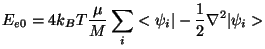 |
|
|
(35.12) |
where 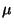 is the ficticious electronic mass,
is the ficticious electronic mass, 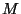 is average mass of one atom,
and
is average mass of one atom,
and
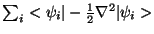 is the kinetic energy of the
electrons.
is the kinetic energy of the
electrons.
Blöchl and Parrinello suggested that the choice of mass parameters,
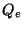 , and
, and 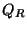 should be made such that the period of oscillating thermostats
should be chosen larger than the typical time scale for the dynamical events of
interest but shorter than the simulation time.
should be made such that the period of oscillating thermostats
should be chosen larger than the typical time scale for the dynamical events of
interest but shorter than the simulation time.
where 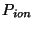 and
and 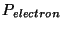 are the periods of oscillation for the ionic and ficatious
electronic thermostats.
are the periods of oscillation for the ionic and ficatious
electronic thermostats.
In simulated annealing simulations the electronic and ionic Temperatures are scaled
according to an exponential cooling schedule,
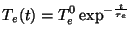 |
|
|
(35.15) |
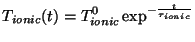 |
|
|
(35.16) |
where 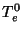 and
and 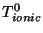 are the intial temperatures, and
are the intial temperatures, and 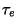 and
and 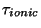 are the cooling rates in atomic units.
are the cooling rates in atomic units.
35.7 PSPW Tutorial 1: Minimizing the geometry for a C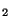 molecule
molecule
In this section we show how use the PSPW module to optimize the geometry
for a C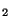 molecule at the PBE96 levels.
molecule at the PBE96 levels.
In the following example we show the input needed to optimize the geometry
for a C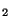 molecule at the LDA level. In this example, default pseudopotentials
from the pseudopotential library are used for C, the boundary condition is free-space,
the exchange correlation functional is PBE96, The boundary condition is free-space, and
the simulation cell cell is aperiodic and cubic with a side length of 10.0 Angstroms and has
40 grid points in each direction (cutoff energy is 44 Ry).
molecule at the LDA level. In this example, default pseudopotentials
from the pseudopotential library are used for C, the boundary condition is free-space,
the exchange correlation functional is PBE96, The boundary condition is free-space, and
the simulation cell cell is aperiodic and cubic with a side length of 10.0 Angstroms and has
40 grid points in each direction (cutoff energy is 44 Ry).
start c2_pspw_pbe96
title "C2 restricted singlet dimer optimization - PBE96/44Ry"
geometry
C -0.62 0.0 0.0
C 0.62 0.0 0.0
end
pspw
simulation_cell units angstroms
boundary_conditions aperiodic
SC 10.0
ngrid 40 40 40
end
xc pbe96
end
set nwpw:minimizer 2
task pspw optimize
35.8 PSPW Tutorial 2: Running a Car-Parrinello Simulation
In this section we show how use the PSPW module to perform a Car-Parrinello
molecular dynamic simulation for a C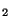 molecule at the LDA level.
Before running a PSPW Car-Parrinello simulation the system should be
on the Born-Oppenheimer surface, i.e. the one-electron orbitals should be minimized
with respect to the total energy (i.e. task pspw energy). The input needed
is basically the same as for optimizing the geometry of a C
molecule at the LDA level.
Before running a PSPW Car-Parrinello simulation the system should be
on the Born-Oppenheimer surface, i.e. the one-electron orbitals should be minimized
with respect to the total energy (i.e. task pspw energy). The input needed
is basically the same as for optimizing the geometry of a C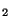 molecule at the LDA level,
except that and additional Car-Parrinello sub-block is added.
molecule at the LDA level,
except that and additional Car-Parrinello sub-block is added.
In the following example we show the input needed to run a Car-Parrinello simulation
for a C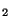 molecule at the LDA level. In this example, default pseudopotentials
from the pseudopotential library are used for C, the boundary condition is free-space,
the exchange correlation functional is LDA, The boundary condition is free-space, and
the simulation cell cell is aperiodic and cubic with a side length of 10.0 Angstroms and has
40 grid points in each direction (cutoff energy is 44 Ry). The time step and fake mass
for the Car-Parrinello run are specified to be 5.0 au and 600.0 au, respectively.
molecule at the LDA level. In this example, default pseudopotentials
from the pseudopotential library are used for C, the boundary condition is free-space,
the exchange correlation functional is LDA, The boundary condition is free-space, and
the simulation cell cell is aperiodic and cubic with a side length of 10.0 Angstroms and has
40 grid points in each direction (cutoff energy is 44 Ry). The time step and fake mass
for the Car-Parrinello run are specified to be 5.0 au and 600.0 au, respectively.
start c2_pspw_lda_md
title "C2 restricted singlet dimer, LDA/44Ry - constant energy Car-Parrinello simulation"
geometry
C -0.62 0.0 0.0
C 0.62 0.0 0.0
end
pspw
simulation_cell units angstroms
boundary_conditions aperiodic
lattice
lat_a 10.00d0
lat_b 10.00d0
lat_c 10.00d0
end
ngrid 40 40 40
end
Car-Parrinello
fake_mass 600.0
time_step 5.0
loop 10 10
end
end
set nwpw:minimizer 2
task pspw energy
task pspw Car-Parrinello
35.9 PSPW Tutorial 3: Running a Simulated Annealing Car-Parrinello Simulation
35.10 PSPW Tutorial 4: optimizing a unit cell and geometry for Silicon-Carbide
The following example demonstrates how to uses the PSPW module to optimize the unit cell
and geometry for a silicon-carbide crystal.
title "SiC 8 atom cubic cell - geometry and unit cell optimization"
start SiC
#**** Enter the geometry using fractional coordinates ****
geometry units au center noautosym noautoz print
system crystal
lat_a 8.277d0
lat_b 8.277d0
lat_c 8.277d0
alpha 90.0d0
beta 90.0d0
gamma 90.0d0
end
Si -0.50000d0 -0.50000d0 -0.50000d0
Si 0.00000d0 0.00000d0 -0.50000d0
Si 0.00000d0 -0.50000d0 0.00000d0
Si -0.50000d0 0.00000d0 0.00000d0
C -0.25000d0 -0.25000d0 -0.25000d0
C 0.25000d0 0.25000d0 -0.25000d0
C 0.25000d0 -0.25000d0 0.25000d0
C -0.25000d0 0.25000d0 0.25000d0
end
#***** setup the nwpw gamma point code ****
nwpw
simulation_cell
ngrid 16 16 16
end
ewald_ncut 8
end
set nwpw:minimizer 2
set nwpw:psi_nolattice .true. # turns of unit cell checking for wavefunctions
driver
clear
maxiter 40
end
set includestress .true. # this option tells driver to optimize the unit cell
task pspw optimize
35.11 Band Tutorial 1: Minimizing the energy of a silicon-carbide crystal by running a PSPW and Band simulation in tandem
The following input deck performs a PSPW energy calculation followed
by a Band energy calculation at the 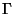 -point for a cubic (8-atom)
silicon-carbide crystal. Since the geometry is entered using fractional coordinates
the unit cell parameters do not have to be re-specified in the simulation_cell
nwpw sub-block. In this example, default pseudopotential from the pseudopotential
library are used for C and Si. The advantage of running these calculations in tandem is that
the Band code uses the wavefunctions generated from the faster PSPW calculation for
its initial guess. The PSPW energy is -38.353570, and the Band energy is -38.353570.
-point for a cubic (8-atom)
silicon-carbide crystal. Since the geometry is entered using fractional coordinates
the unit cell parameters do not have to be re-specified in the simulation_cell
nwpw sub-block. In this example, default pseudopotential from the pseudopotential
library are used for C and Si. The advantage of running these calculations in tandem is that
the Band code uses the wavefunctions generated from the faster PSPW calculation for
its initial guess. The PSPW energy is -38.353570, and the Band energy is -38.353570.
start SiC_band
title "SiC 8 atom cubic cell"
#**** geometry entered using fractional coordinates ****
geometry units au center noautosym noautoz print
system crystal
lat_a 8.277d0
lat_b 8.277d0
lat_c 8.277d0
alpha 90.0d0
beta 90.0d0
gamma 90.0d0
end
Si -0.50000d0 -0.50000d0 -0.50000d0
Si 0.00000d0 0.00000d0 -0.50000d0
Si 0.00000d0 -0.50000d0 0.00000d0
Si -0.50000d0 0.00000d0 0.00000d0
C -0.25000d0 -0.25000d0 -0.25000d0
C 0.25000d0 0.25000d0 -0.25000d0
C 0.25000d0 -0.25000d0 0.25000d0
C -0.25000d0 0.25000d0 0.25000d0
end
#***** setup the nwpw gamma point code ****
nwpw
simulation_cell
ngrid 16 16 16
end
brillouin_zone
kvector 0.0 0.0 0.0
end
ewald_ncut 8
end
set nwpw:minimizer 2
set nwpw:psi_brillioun_check .false.
task pspw energy
task band energy
35.12 PAW Tutorial
The following input deck performs for a water molecule a PSPW energy calculation followed
by a PAW energy calculation and a PAW geometry optimization calculation.
The default unit cell paramters are used (SC=20.0, ngrid 32 32 32). In this simulation, the
first PAW run optimizes the wavefunction and the second PAW run optimizes the wavefunction
and geometry in tandem.
title "paw steepest descent test"
start paw_test
charge 0
geometry units au nocenter noautoz noautosym
O 0.00000 0.00000 0.01390
H -1.49490 0.00000 -1.18710
H 1.49490 0.00000 -1.18710
end
nwpw
time_step 15.8
ewald_rcut 1.50
tolerances 1.0d-8 1.0d-8
end
set nwpw:lcao_iterations 1
set nwpw:minimizer 2
task pspw energy
task paw steepest_descent
nwpw
time_step 5.8
geometry_optimize
ewald_rcut 1.50
tolerances 1.0d-7 1.0d-7 1.0d-4
end
task paw steepest_descent
35.13 NWPW Capabilities and Limitations
- You cannot use more processors than the size of the third dimension
(e.g. a 64x64x64 FFT grid can use at most 64 processors).
- The second and third dimensions of the FFT grid must be the same
(i.e. the parameters na2 and na3 must be the same for each simulation cell).
- Wannier orbital generation only works with cubic unit cells (
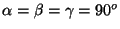 )
)
- A default Brillouin zone has not been defined for Band simulations.
- PAW only works with units cells that have periodic boundary conditions.
- PAW is not interfaced to driver, stepper, and vib modules.
Questions and encountered problems should be reported to
nwchem-support@emsl.pnl.gov
or to Eric J. Bylaska, Eric.Bylaska@pnl.gov



Next: 36. Controlling NWChem with
Up: user
Previous: 34. File formats
Contents
2003-10-08
![[*]](crossref.png)
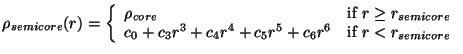
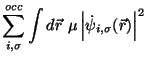
![$\displaystyle \sum_{i,\sigma}^{occ} \int d\vec{r}\ \mu \left\vert
\dot{\psi}_{i...
...eft[ \left\{ \psi_{i,\sigma}(\vec{r})\right\},\left\{\vec{R}_I \right\} \right]$](img286.gif)
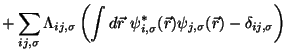
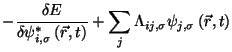
![$\displaystyle 2 \psi_{i,\sigma}^{t} - \psi_{i,\sigma}^{t-\Delta t}
+ \frac{(\De...
...\psi_{i,\sigma}^{*}}
+ \sum_{j} \psi_{j,\sigma} \Lambda_{ji,\sigma}
\right]_{t}$](img298.gif)