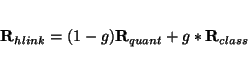


Next: 34. File formats
Up: user
Previous: 32. Analysis
Contents
Subsections
Combined or hybrid Quantum Mechanics and Molecular Mechanics (QM/MM)
is a simulation methodology 33.1 that is about 15 years old but in all the
literature there are cautions that calibration computations must be
done to validate the model for each particular chemical system
studied. This is not a black box style computation and the NWChem
users are advised that without calibration QM/MM may not give the
appropriate results. Since both quantum-mechanical and classical
molecular mechanics are involved in the calculation good working knowledge of the two methods
is required to ensure meaningful results.
The QM/MM module is invoked with the following task directive.
task qmmm <string qmtheory> <string operation> [numerical] [ignore]
where qmtheory specifies quantum method for the calculation of the quantum region. It is expected that
most of the QM/MM simulations will be performed with
HF or DFT theories, but any other QM theory supported by NWChem should work as well.
Currently the supported operations for QM/MM runs
are energy, optimize, saddle, dynamics, numerical hessian, and numerical
frequencies.
Unlike pure
quantum mechanical calculations the information about the chemical system
for QM/MM simulations is contained not in the geometry block but in the externally prepared topology and restart files.
These files have to be present prior to any QM/MM simulation.
The input file for QM/MM simulation can be divided into three major parts - specification of the molecular
mechanics parameters for the classical region, specification of the quantum mechanical method for the quantum region,
and the parameters of the interaction between quantum and classical methods.
All this discussed in detail in the sections below.
Generated by the prepare
module (see section 30) restart and topology files contain
information about the classical force field as well as the
coordinates of quantum (qm) and molecular
mechanics (mm) regions.
In a typical setting this "preparation stage"
will be run separately from main QM/MM simulation. This will require a
properly formatted
PDB file for the system. In more complex cases (e.g.non-standard residues or nucleotides) additional fragment and parameter
files might have to be provided by the user. The definition of the quantum region
in the input for the prepare module is specified by either modify atom directive (see Section 30):
modify atom <integer isgm>:<string atomname> quantum
or modify segment directive
modify segment <integer isgm> quantum
Here isgm and atomname refer to the residue number and atom name record
as given in the PDB file.
It is important to note that
that the leading blanks
in atom name record should be indicated with underscores.
Per PDB format quidelines the atom name record starts at column 13. If, for example,
the atom name record "OW" starts
in the 14th column in PDB file, it will appear
as "_OW" in the modify atom directive in the prepare block. In the current implementation
only solute atoms can be declared as quantum. If part of the solvent has to be treated quantum mechanically
then it has to redeclared to be solute.
In addition to modify commands the prepare input block should
also contain update lists and ignore directives. There are other options
that can be used in the input block for the prepare module ( e.g. solvating the structure, etc ),
those discussed
in more details in Section 30.
The successful run of the prepare module will result in generation of
topology and restart files. Similar to classical MD, both files are required for QM/MM simulations
and have to be placed in the same directory
as the input file. Here is an example input file that will generate QM/MM restart and topology files for the ethanol molecule
title "Prepare QM/MM calculation of ethanol"
start etl
prepare
#--name of the pdb file
source etl0.pdb
#--generate new topology and sequence file
new_top new_seq
#--generate new restart file
new_rst
#--define quantum region (note the use of underscore)
modify atom 1:_C1 quantum
modify atom 1:2H1 quantum
modify atom 1:3H1 quantum
modify atom 1:4H1 quantum
#
update lists
ignore
end
task prepare
These are contents of etl0.pdb file used in the above input file.
ATOM 1 O etl 1 1.201 -0.271 -0.000 1.00 0.00 O
ATOM 2 H etl 1 1.995 0.329 -0.000 1.00 0.00 H
ATOM 3 C1 etl 1 -1.180 -0.393 0.000 1.00 0.00 C
ATOM 4 2H1 etl 1 -2.128 0.155 -0.000 1.00 0.00 H
ATOM 5 3H1 etl 1 -1.130 -1.030 0.887 1.00 0.00 H
ATOM 6 4H1 etl 1 -1.130 -1.030 -0.887 1.00 0.00 H
ATOM 7 C2 etl 1 0.006 0.573 0.000 1.00 0.00 C
ATOM 8 2H2 etl 1 -0.042 1.220 0.890 1.00 0.00 H
ATOM 9 3H2 etl 1 -0.042 1.220 -0.890 1.00 0.00 H
END
Running the input shown above will produce (among other things) the topology file (etl.top) and the restart file
(etl_md.rst). The naming of the topology file follows after the rtdb name specified in the start directive in the input (i.e. "start etl"),
while the "_md" suffix in the restart file name is specific to the way prepare module works in this particular case. If necessary, this
particular naming scheme can be altered using system keyword in the prepare input block (for more details see Section 30).
The molecular mechanics parameters are given in the form of standard MD input block as
used by the MD module (c.f. Section 31). This input block
is required for QM/MM simulations. It specifies the
restart and topology file that will be used in the calculation.
It also contains information relevant to the calculation
of the classical region
(e.g. cutoff distances, constraints, optimization and dynamics parameters, etc)
in the system. This input block may also contain fixed atom constraints on both classical and quantum atoms. Continuing with our
example for ethanol molecule here is a simple MD input block that may be used for this system.
md
# this specifies that etl_md.rst will be used as a restart file
# and etl.top will be a topology file
system etl_md
# if we ever wanted to fix C1 atom
fix solute 1 _C1
end
The parameters defining calculation of the QM region (including basis sets)
must be present in the traditional NWChem input format
except for the geometry block. The geometrical information will
be constructed automatically by QM/MM using information from the MD module.
The QM/MM interface parameters define the interaction between classical and quantum regions.
The input follows standard NWChem format:
qmmm
[ eref <double precision default 0.0d0>]
[ bqzone <double precision default 9.0d0>]
[ bq_exclude <(none||all||linkbond||linkbond_H) default none>]
[ bq_update <(static||dynamic) >]
[ link_atoms <(hydrogen||halogen) default hydrogen>]
[ link_ecp <(auto||user) default auto>]
[ optimization <(all||mm|qm) default qm>]
end
Detailed explanation of the subdirectives in the QM/MM input block is given below:
eref <double precision default 0.0d0>
This directive sets the relative zero of energy for the
QM component of the system. The need for this directive
arizes from different definitions of zero energy for QM and MM methods.
Most QM methods define the zero of energy for the system as
vacuum. The zero of energy for the MM system is by
definition of most parameterized force fields the separated atom
energy. Therefore in many cases the energetics of the QM system
will likely overshadow the
MM component of the system. This imbalance can be corrected by
suitably chosen value of eref
bqzone <double precision default 9.0d0>
This directive defines the radius of the zone (in angstroms) around the quantum region
where classical residues/segments
will be allowed to interact with quantum region electrostatically. It should be noted
that classical atoms interacting with quantum region via bonded interactions are always
included in the bqzone (this is true even if bqzone is set to 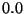 ). In addition, even if one atom
of a given charged group is in the bqzone (residues are typically treated as one charged group) then the whole
group will be included in the bqzone.
). In addition, even if one atom
of a given charged group is in the bqzone (residues are typically treated as one charged group) then the whole
group will be included in the bqzone.
bq_exclude <(none||all||linkbond||linkbond_H) default none>
This directive operates in conjunction with bqzone keyword offering additional level of control
on electrostatic interactions between quantum and classical regions. Default value none will
leave bqzone unchanged, all will remove all
electrostatic interactions between
classical and quantum regions,
linkbond will result in the removal of electrostatic interactions with atoms that are connected to a quantum region
by at most two bonds,
linkbond_H is similar to linkbond but will only remove interactions with hydrogen atoms.
If necccessary, further modifications of the electrostatic interactions can be achieved
using the prepare module30.
bq_update <(static||dynamic)
This directive controls whether the bqzone stays fixed (static) or constantly
updated (dynamic) during the calculation. In most cases direct specification of this keyword
will not be necessary as its value will be set automatically based on the nature of the calculation.
link_atoms <(hydrogen||halogen) default halogen>
This directive controls the treatment of bonds crossing the boundary between quantum and classical regions.
The use of hydrogen keyword will trigger truncation of such bonds with hydrogen link atoms. The position of the hydrogen
atom will be calculated from the coordinates of the quantum and classical atom of the truncated bond using the
following expression
where 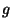 is the scale factor set at
is the scale factor set at 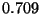
Setting link_atoms to halogen will result in the modification of the quantum
atom of the truncated bond to
to the fluoride atom. This fluoride atom will typically carry an effective core potential (ECP) basis set as specified
in link_ecp directive.
link_ecp <(auto||user) default auto>
This directive specifies ECP basis set on fluoride link atoms. If set to auto
the ECP basis set given by Zhang, Lee, Yang for 6-31G* basis.33.2
will be used. Strictly speaking, this implies the use of 6-31G* spherical basis as the main basis set.
If other choices are desired then keyword user should be used and ECP basis set should be entered separatelly
following the format given in
section 8.
The name tag for fluoride link atoms is F_L.
optimization <(all||mm|qm) default qm>
This directive specifies which region will be optimized during QM/MM optimization. If set to all both qm and mm
region will be optimized simultaneously using mm optimization module, if mm only mm portion will be optimized
using mm optimization module, finally if qm is specified then only qm region will be optimized using driver
module. Since option all is based on steepest descent minimization algorithm it is not reccomended for
accurate QM/MM optimization tasks. The most efficient and accurate way to perform QM/MM optimizations is
to optimize quantum region using optimization qm followed by the relaxation of the mm region using optimization mm.



Next: 34. File formats
Up: user
Previous: 32. Analysis
Contents
2005-09-12
![]() ). In addition, even if one atom
of a given charged group is in the bqzone (residues are typically treated as one charged group) then the whole
group will be included in the bqzone.
). In addition, even if one atom
of a given charged group is in the bqzone (residues are typically treated as one charged group) then the whole
group will be included in the bqzone.